Section 13 Non-parametric tests
Prerequisites
Load the libraries with R:
## Warning: 程辑包'BSDA'是用R版本4.3.3 来建造的## 载入需要的程辑包:lattice##
## 载入程辑包:'BSDA'## The following object is masked from 'package:datasets':
##
## Orange## Warning: 程辑包'FSA'是用R版本4.3.3 来建造的## Registered S3 methods overwritten by 'FSA':
## method from
## confint.boot car
## hist.boot car## ## FSA v0.9.5. See citation('FSA') if used in publication.
## ## Run fishR() for related website and fishR('IFAR') for related book.Section example: Soil organic matter
Given the following organic matter contents (unit: %) in different soil types, do organic matters differ among soil types?
| Soil type | Soil organic matter content (%) |
|---|---|
| A | 2.0, 2.8, 3.3, 3.2, 4.4, 3.6, 1.9, 3.3, 2.8, 1.1 |
| B | 3.5, 2.8, 3.2, 3.5, 2.3, 2.4, 2.0, 1.6 |
| C | 3.3, 3.6, 2.6, 3.1, 3.2, 3.3, 2.9, 3.4, 3.2, 3.2 |
| D | 3.2, 3.3, 3.2, 2.9, 3.3, 2.5, 2.6, 2.8 |
| E | 2.6, 2.6, 2.9, 2.0, 2.0, 2.1 |
| F | 3.1, 2.9, 3.1, 2.5 |
| G | 2.6, 2.2, 2.2, 2.5, 1.2, 1.2 |
| H | 2.5, 2.4, 3.0, 1.5 |
Overview of non-parametric tests
A non-parametric test, also known as a distribution-free test, assumes nothing about the underlying distribution (for example, that the data comes from a normal distribution). That’s compared to parametric test, which makes assumptions about a population’s parameters (for example, the mean or standard deviation); When the word “non-parametric” is used in statistics, it doesn’t quite mean that you know nothing about the population. It usually means that you know the population data does not have a normal distribution.
Situations to use non-parametric tests:
The underlying distribution is not normal, thus the normality assumption is not valid
Data type is nominal (e.g.,
passorfail) or ordinal (aka, ranks or start-rating)One or more assumptions of a parametric test have been violated
The sample size is too small (less than
6) to run a parametric testOutliers can not be removed
Prefer to test for the median rather than the mean (e.g., very skewed distribution)
Non-parametric tests have lower power compared to parametric tests - they often do not reject H0 when they should.
Mann-Whitney U test
The Mann–Whitney U test is also called the Mann–Whitney–Wilcoxon
(MWW), Wilcoxon rank-sum test, or Wilcoxon–Mann–Whitney test. It is used
to compare the differences between two independent samples when the
sample distributions are not normally distributed and the sample sizes
are small (n < 30). It is considered to be the
non-parametric equivalent to the two-sample independent t-test.
The requirements of using Mann–Whitney U test are:
Random and independent samples
For maximum accuracy, there better be no ties. But there is a way to handle ties in the test.
In the Mann–Whitney U test, we have:
H0: There is no difference in the two population means
H1: There is a difference in the two population means
The Mann–Whitney U test is done with the wilcox.test()
function in R:
# Make up samples
Treat <- c(3, 5, 1, 4, 3, 5)
Control <- c(4, 8, 6, 2, 1, 9)
# Perform the Mann Whitney U test
wilcox.test(Treat, Control, paired=F, alternative="two.sided")## Warning in wilcox.test.default(Treat, Control, paired = F, alternative = "two.sided"): 无法精確計算带连结的p值##
## Wilcoxon rank sum test with continuity correction
##
## data: Treat and Control
## W = 13, p-value = 0.468
## alternative hypothesis: true location shift is not equal to 0Wilcoxon signed rank test
The Wilcoxon signed rank test is a non-parametric alternative to paired t-test used to compare paired data. It’s used whendata are not normally distributed.
The requirements of using the Wilcoxon signed rank test are:
Random and independent samples
Approximately symmetrical distributions
In the Wilcoxon signed rank test, we have:
H0: There is no difference in the two population means
H1: There is a difference in the two population means
The Wilcoxon signed rank test is also done with the
wilcox.test() function in R, but with different
keywords:
# Make up samples
Treat <- c(3, 5, 1, 4, 3, 5)
Control <- c(4, 8, 6, 2, 1, 9)
# Perform the Wilcoxon signed rank test
wilcox.test(Treat, Control, paired=T, alternative="two.sided")## Warning in wilcox.test.default(Treat, Control, paired = T, alternative = "two.sided"): 无法精確計算带连结的p值##
## Wilcoxon signed rank test with continuity correction
##
## data: Treat and Control
## V = 5, p-value = 0.2932
## alternative hypothesis: true location shift is not equal to 0Sign test
When the distribution of differences between paired data values is
neither normal nor symmetrical, use the sign test. The sign test is done
with the SIGN.test() function from the BSDA
package R:
##
## Dependent-samples Sign-Test
##
## data: Treat and Control
## S = 2, p-value = 0.6875
## alternative hypothesis: true median difference is not equal to 0
## 95 percent confidence interval:
## -4.9 2.0
## sample estimates:
## median of x-y
## -2
##
## Achieved and Interpolated Confidence Intervals:
##
## Conf.Level L.E.pt U.E.pt
## Lower Achieved CI 0.7812 -4.0 2
## Interpolated CI 0.9500 -4.9 2
## Upper Achieved CI 0.9688 -5.0 2Kruskal–Wallis test
The Kruskal–Wallis test, also known as Kruskal–Wallis H test or one-way ANOVA on ranks, is an alternative for the one-way analysis of variance (ANOVA). The test is for checking whether samples originate from the same distribution. It is used for comparing two or more independent samples. It extends the Mann–Whitney U test, which is used for comparing only two groups.
The test assumes that the observations are independent. That is, it is not appropriate for paired observations or repeated measures data.
In the Kruskal–Wallis test, we have:
H0: There is no difference among population means
H1: There is a difference among population means
The Kruskal–Wallis test is done with the kruskal.test()
function in R:
# Load data
require(graphics)
# Boxplots
ggplot(airquality, aes(x=Month, y=Ozone, group=Month, fill=Month)) +
geom_boxplot() +
labs(title="Monthly ozone",
x="Month", y="Ozone [ppb]") +
theme_classic()## Warning: Removed 37 rows containing non-finite values (`stat_boxplot()`).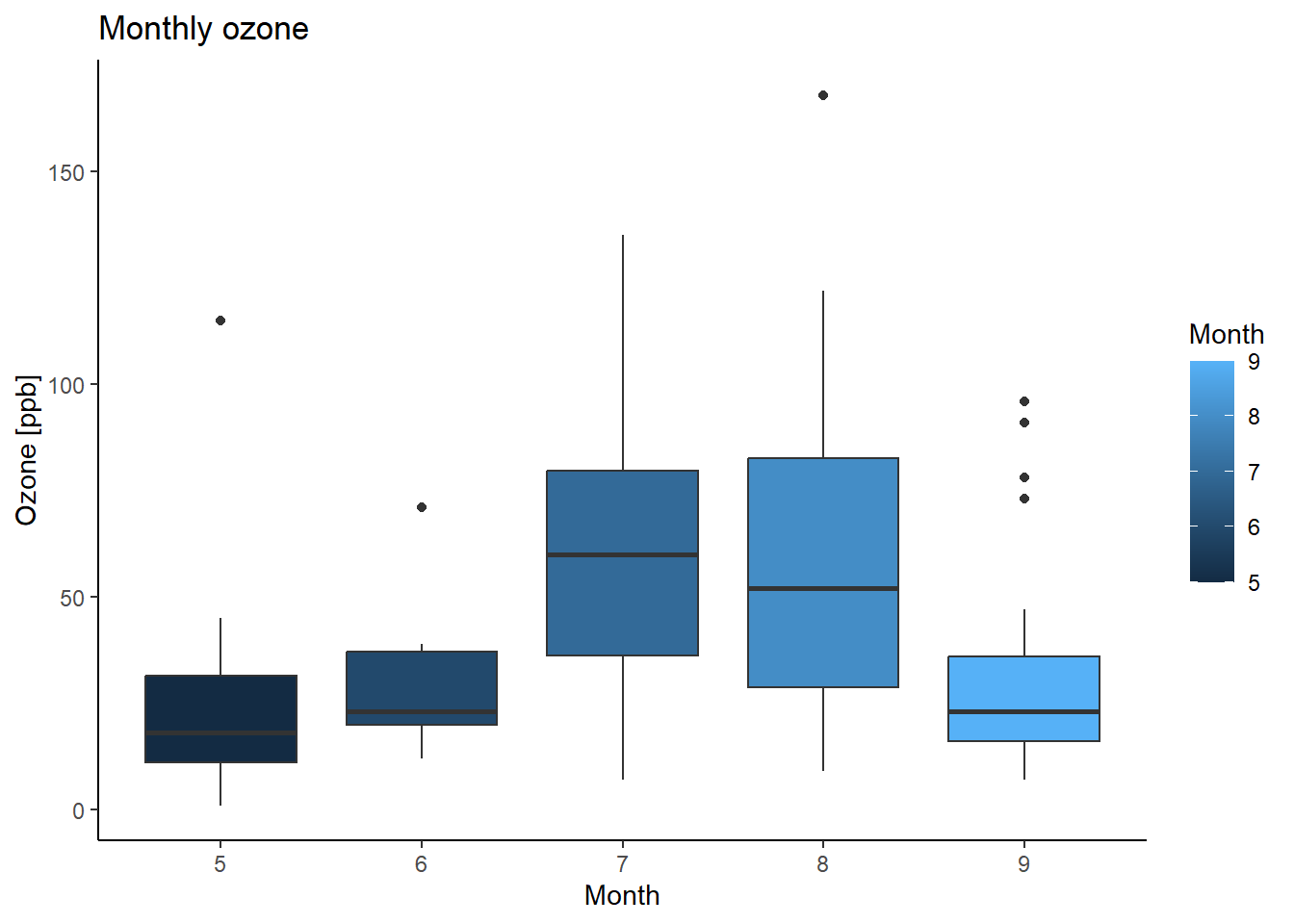
##
## Kruskal-Wallis rank sum test
##
## data: Ozone by Month
## Kruskal-Wallis chi-squared = 29.267, df = 4, p-value = 6.901e-06The outcome of the Kruskal–Wallis test tells you if there are
differences among the group means, but doesn’t tell you which groups are
different from other groups. To determine which groups are different
from others, post-hoc testing can be conducted. The most common post-hoc
test for the Kruskal–Wallis test is the Dunn test, here
conducted with the dunnTest() function in the
FSA package.
## Warning: Month was coerced to a factor.## Warning: Some rows deleted from 'x' and 'g' because missing data.## Dunn (1964) Kruskal-Wallis multiple comparison## p-values adjusted with the Benjamini-Hochberg method.## Comparison Z P.unadj P.adj
## 1 5 - 6 -0.925158616 3.548834e-01 4.436043e-01
## 2 5 - 7 -4.419470641 9.894296e-06 9.894296e-05
## 3 6 - 7 -2.244208032 2.481902e-02 4.963804e-02
## 4 5 - 8 -4.132813422 3.583496e-05 1.791748e-04
## 5 6 - 8 -2.038635487 4.148642e-02 6.914403e-02
## 6 7 - 8 0.286657218 7.743748e-01 8.604164e-01
## 7 5 - 9 -1.321202283 1.864339e-01 2.663342e-01
## 8 6 - 9 0.002538555 9.979745e-01 9.979745e-01
## 9 7 - 9 3.217199124 1.294487e-03 4.314957e-03
## 10 8 - 9 2.922827778 3.468683e-03 8.671708e-03Levene test
The Levene test checks whether several groups have the same variance in the population (homoscedasticity). Levene test is therefore used to test the null hypothesis that the samples to be compared come from a population with the same variance.
- H0: The samples are from populations with equal variances
- H1: At least one population variance is not equal to the others
A big advantage of Levene test is that it is very stable against violations of the normal distribution. This makes Levene test is used in many statistics programs.
# Sub-samples
Control <- c(0.64, 0.91, 0.84, 0.41, 1.58, 0.48, 0.88, 0.74, 1.09, 1.28)
P1 <- c(1.20, 1.17, 0.76, 0.92, 0.65, 1.14, 1.05, 1.28, 0.89, 1.06)
P2 <- c(0.25, 1.31, 0.43, 1.18, 1.02, 0.83, 1.02, 0.66, 0.70, 0.83)
P3 <- c(0.63, 0.30, 0.49, 0.01, 0.63, 0.69, 0.68, 0.34, 1.09, 0.42)
# Make data frame
Cd_data <- data.frame(Cd=c(Control, P1, P2, P3),
group=c(rep("Control", length(Control)),
rep("Plant 1", length(P1)),
rep("Plant 2", length(P2)),
rep("Plant 3", length(P3))) )
# Change to factor type using tibble
cd_data_tbl <- as_tibble(Cd_data) %>%
mutate(group = factor(group, ordered = TRUE))
# Test homogeneity of variances
require(car)## 载入需要的程辑包:car## Warning: 程辑包'car'是用R版本4.3.2 来建造的## 载入需要的程辑包:carData## Warning: 程辑包'carData'是用R版本4.3.2 来建造的##
## 载入程辑包:'carData'## The following objects are masked from 'package:BSDA':
##
## Vocab, Wool##
## 载入程辑包:'car'## The following object is masked from 'package:FSA':
##
## bootCase## The following object is masked from 'package:EnvStats':
##
## qqPlot## The following object is masked from 'package:gtools':
##
## logit## The following object is masked from 'package:dplyr':
##
## recode## Levene's Test for Homogeneity of Variance (center = median)
## Df F value Pr(>F)
## group 3 0.6261 0.6029
## 36Spearman correlation test
The Spearman correlation test is a rank-based test that does not
require assumptions about the distribution of the data. The correlation
coefficient from the test, \(\rho\),
ranges from -1 to –1, with +1
being a perfect positive correlation and –1 being a perfect
negative correlation. A \(\rho\) of
0 represents no correlation.
In the Spearman correlation test, we have:
H0: The populations do not correlate with each other
H1: The populations correlate with each other
The Spearman correlation is done with the cor.test()
function in R:
# Make up some random values
x <- rnorm(20,0,1)
y <- 2*x+rnorm(20,0,0.5)
# Perform the Spearman correlation test
cor.test(x, y, method="spearman", alternative="two.sided", conf.level=0.95)##
## Spearman's rank correlation rho
##
## data: x and y
## S = 148, p-value < 2.2e-16
## alternative hypothesis: true rho is not equal to 0
## sample estimates:
## rho
## 0.8887218Kendall correlation test
The Kendall correlation test is also a rank-based test. The
correlation coefficient from the test is \(\tau\), ranging from -1 to
+1.
Used the Kendall correlation when the sample size is small or there are many tied ranks.
In the Kendall correlation test, we have:
H0: The populations do not correlate with each other
H1: The populations correlate with each other
The Kendall correlation is also done with the cor.test()
function in R:
# Perform the Kendall correlation test
cor.test(x, y, method="kendall", alternative="two.sided", conf.level=0.95)##
## Kendall's rank correlation tau
##
## data: x and y
## T = 165, p-value = 5.089e-07
## alternative hypothesis: true tau is not equal to 0
## sample estimates:
## tau
## 0.7368421Summary of non-parametric tests
| Purpose | Parametric | Alternative non-parametric |
|---|---|---|
| Checking independence | Runs test | |
| Checking normality | Shapiro-Wilk test, Lillifors test | |
| Checking Outliers | Grubbs’ test, Dixon’s test, Rosner’s test | Walsh’s test |
| Comparing means of two independent populations | t test | Mann-Whitney U test |
| Comparing means of two paired populations | paired t test | Wilcoxon signed rank, Signed test |
| Comparing means of more than 2 independent populations | One-way ANOVA | Kruskal–Wallis test |
| Checking homogeneity of variances | Bartlett test | Levene Test |
| Checking correlation | Pearson test | Spearman test, Kendall test |
In-class exercises
Exercise #1
Using data from the section example, do organic matters differ among soil types? Report your results.